From Command Line¶
Analyze your dataset¶
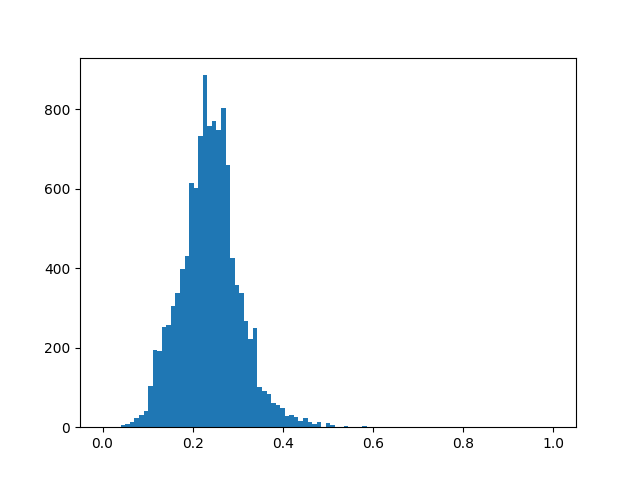
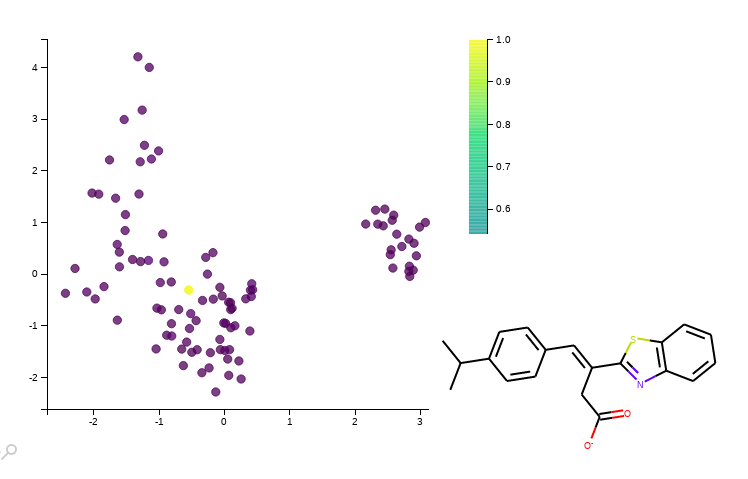
The command below will output the tanimoto similarity distribution among all dataset and all fingerprints, at the same time will show a plot of the two first components of the PCA over the fingerprint space coloured by similarity to your query molecule. If we hover the points of the plot we can inspect the different structures of the molecules.
python -m analogs_finder.main <database> <molecule_query> --analysis
#Use uniform manifold to plot the chemical space
python -m analogs_finder.main analogs_finder/examples/database.sdf analogs_finder/examples/substructre_1.sdf --analysis --dim_type umap
#Use pca to plot the chemical space
python -m analogs_finder.main analogs_finder/examples/database.sdf analogs_finder/examples/substructre_1.sdf --analysis --dim_type pca
We find the similarity_hist_DL.png:
And a firefox window opens retrieving and interactive plot:
N most similar structures¶
Given a database.sdf and a molecule.sdf will output the n most similar structures to the query molecule (–n_structs to specify the number of outputted structures)
python -m analogs_finder.mainpy <database (sdf)> <query_molecule (sdf)> --sb --n <number of output structs> --output <outputname> python -m analogs_finder.main analogs_finder/examples/database.sdf analogs_finder/examples/substructre_1.sdf --sb --n 20 --output most_similars.sdf
Tanimoto similarity search¶
Will retrieve all molecules on the database.sdf that has a tanimoto similarity higher than treshold in respect to the query molecule (0.7 by default, you can specify by –tresh 0.6)
python -m analogs_finder.main <database (sdf)> <query_molecule (sdf)> --treshold tanimoto_treshold --output <outputname> python -m analogs_finder.main analogs_finder/examples/database.sdf analogs_finder/examples/substructre_1.sdf --output most_similars.sdf --tresh 0.7
Search for one or more substructure¶
For each entry in database.py search for the substructure/s present in the query_molecule.sdf
python -m analogs_finder.main <database (sdf)> <substructure_moleculeS (sdf with several entries (substructures)> --substructure --output <outputname> python -m analogs_finder.main analogs_finder/examples/database.sdf analogs_finder/examples/substructre_1.sdf --output most_similars.sdf --substructure
Search for at least one of the substructures in each sdf file¶
For each entry in database.py search for at least one substructure present in the each sdf file
python -m analogs_finder.main <database (sdf)> <substructure_moleculeS (sdf with several entries (substructures)> --combi_subsearch --output <outputname> python -m analogs_finder.main analogs_finder/examples/database.sdf analogs_finder/examples/substructre_*.sdf --output most_similars.sdf --combi_subsearch
Search for similarity and substructure¶
For each entry in database.py search for at least one substructure present in the each sdf file
python -m analogs_finder.main <database (sdf)> <query_molecule (sdf with several entries (substructures)> --output <outputname> --hybrid <substructure sdf file> python -m analogs_finder.main analogs_finder/examples/database.sdf analogs_finder/examples/substructure_2.sdf --output most_similars.sdf --hybrid analogs_finder/examples/substructure_1.sdf
Change fingerprint type¶
python -m analogs_finder.main <database (sdf)> <query_molecule (sdf with several entries (substructures)> --output <outputname> --hybrid <substructure sdf file> --fp_type [ DL, circular, MACCS, torsions, pharm] python -m analogs_finder.main analogs_finder/examples/database.sdf analogs_finder/examples/substructre_2.sdf --output most_similars.sdf --hybrid analogs_finder/examples/substructure_1.sdf --fp_type circular
Use all fingerprints in one job with different tresholds¶
python -m analogs_finder.main <databaseSDF> <querymolecSDF> --tresh <tresholds> --fp_type <fingerpinttypes>
python -m analogs_finder.main ~/repos/analogs_finder/tests/data/database.sdf ~/repos/analogs_finder/tests/data/substructre_1.sdf --tresh 0.7 0.4 0.7 0.27 --fp_type DL circular torsions MACCS
Turbo search method:¶
Instead of just querying the reference molecule and setting a tanimoto treshold, we first look for the N most similar neighbours and we run similarity search with the reference molecule and theses neghbours, finally performing data fusion.
For more details: https://onlinelibrary.wiley.com/doi/abs/10.1002/sam.10037
python -m analogs_finder.main <databaseSDF> <querymolecSDF> --turbo --neighbours <N>--tresh <tresholds> --fp_type <fingerpinttypes>
python -m analogs_finder.main ~/repos/analogs_finder/tests/data/database.sdf ~/repos/analogs_finder/tests/data/substructre_1.sdf --turbo --neighbours 5 --tresh 0.7 --fp_type circular
PostFilter by:¶
To postfilter, add the flags to apply the filters after the method search command. For instance, here we remove duplicates after doing a tanimoto similarity search
python -m analogs_finder.main result_search.sdf substructre_1.sdf --most_similars --remove_duplicates
To postfilter a previously done analog search provide the sdf of the previous analog search as the database followed by -only_postprocess. Here, we remove duplicates of the previous analog serch resut_search.sdf
python -m analogs_finder.main result_search.sdf substructre_1.sdf --only_postfilter --remove_duplicates
Position of growing¶
To only keep the molecules that have a radical growin in a specific initial atom (atom 2 for example) use the command below. To know the indexes of the atoms you can select them with maestro/pymol with the option of labeling atoms by index.
python -m analogs_finder.main result_search.sdf substructre_1.sdf --only_postfilter --atom_to_grow 2
It is also possible to sum them up. For intance, here we keep analogs grown by the atom index 2 or 4
python -m analogs_finder.main result_search.sdf substructre_1.sdf --only_postfilter --atom_to_grow 2 4
To keep analogs that have no radical in a specific atom use –atom_to_avoid. For example, next we keep all molecules that have no radical in the atom index 2 and 4
python -m analogs_finder.main result_search.sdf substructre_1.sdf --only_postfilter --atom_to_avoid 2 4