PELE basis¶
Our Protein Energy Landscape Exploration (PELE) tool combines protein structure prediction algorithms and Metropolis Monte Carlo techniques to efficiently tackle tasks like predicting binding sites, docking pose refinement or modelling exit path of a ligand.
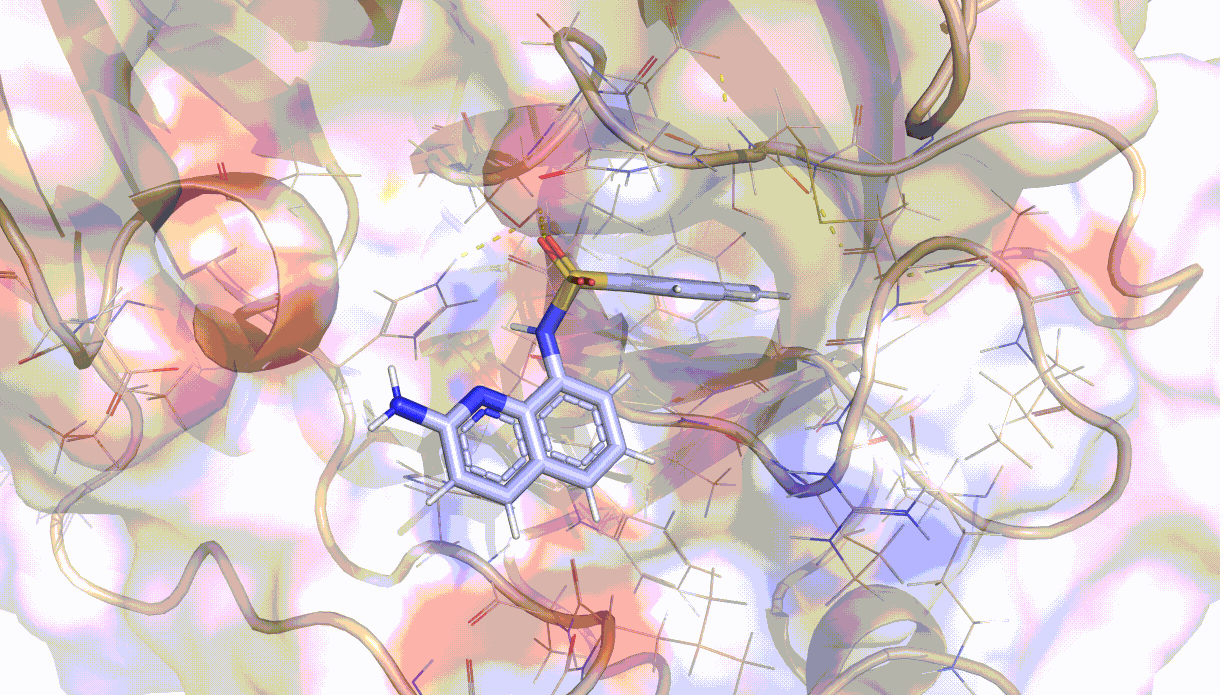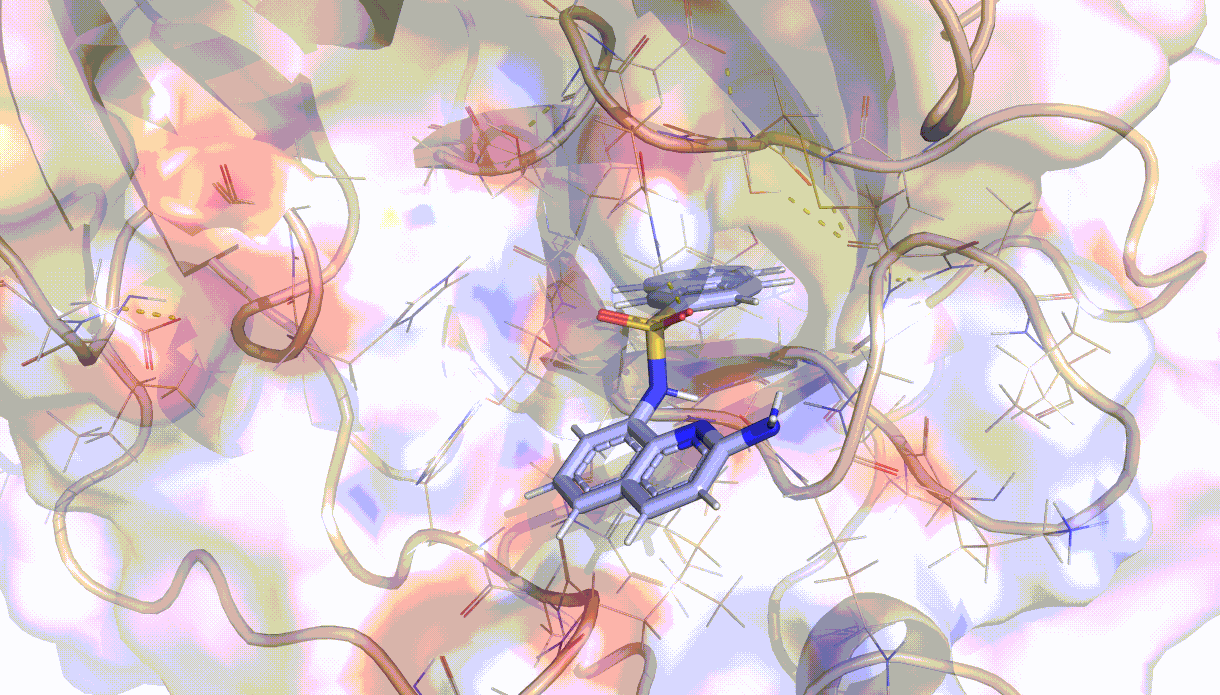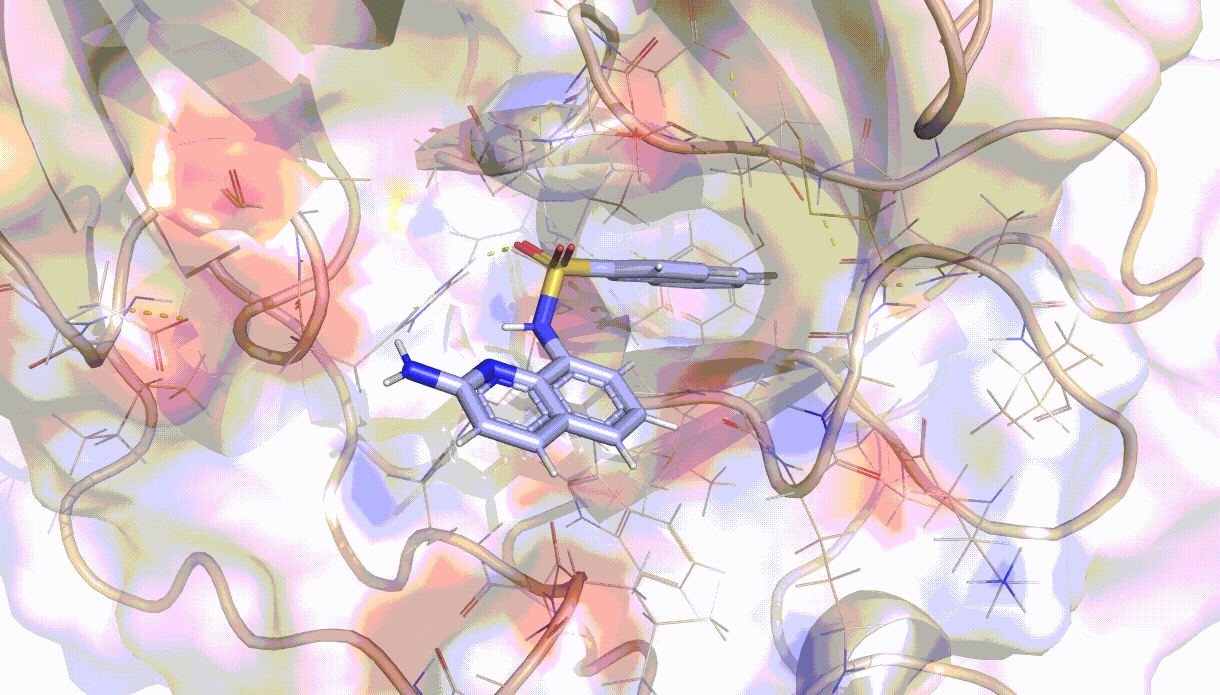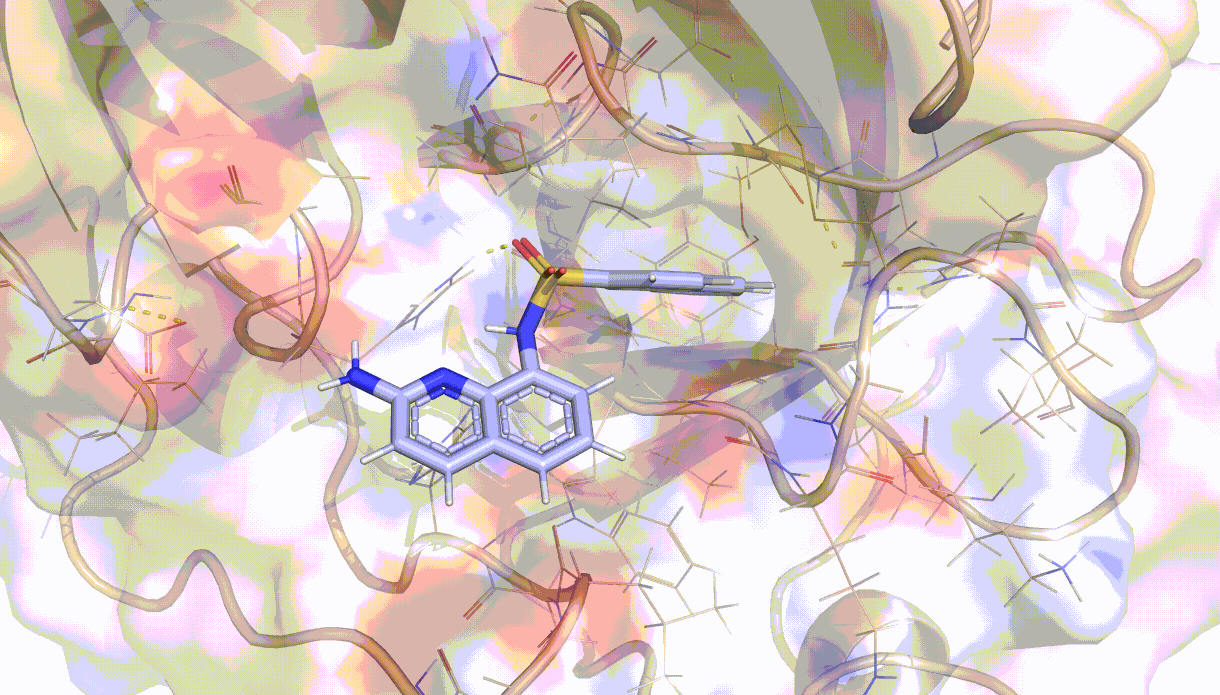
Visualisation of the PELE algorithm¶
If you would like to find out more about the C++ engine running underneath, please refer to the PELE++ documentation.
PELE algorithm¶
Each simulation consists of several steps executing the following algorithm:
1. Perturbation. Localised perturbation of the ligand (if present), involving random translation and rotation, followed by simple side chain relocation to avoid clashes. Additionally, the protein is minimized by driving alpha carbons to new positions resulting from a small displacement in a low frequency anisotropic normal mode (ANM).
➜ This step might also involve water perturbation - find out more about AquaPELE.
2. Relaxation. Optimization of side chains in proximity to the ligand as well as those whose energy changed the most during ANM, using a rotamer library with a resolution of 10°. This is followed by a global minimization with Truncated Newton minimizer.
3. Acceptance. The new structure is accepted (defining a new minimum) or rejected based on the Metropolis criterion.
Check how to set it up here.
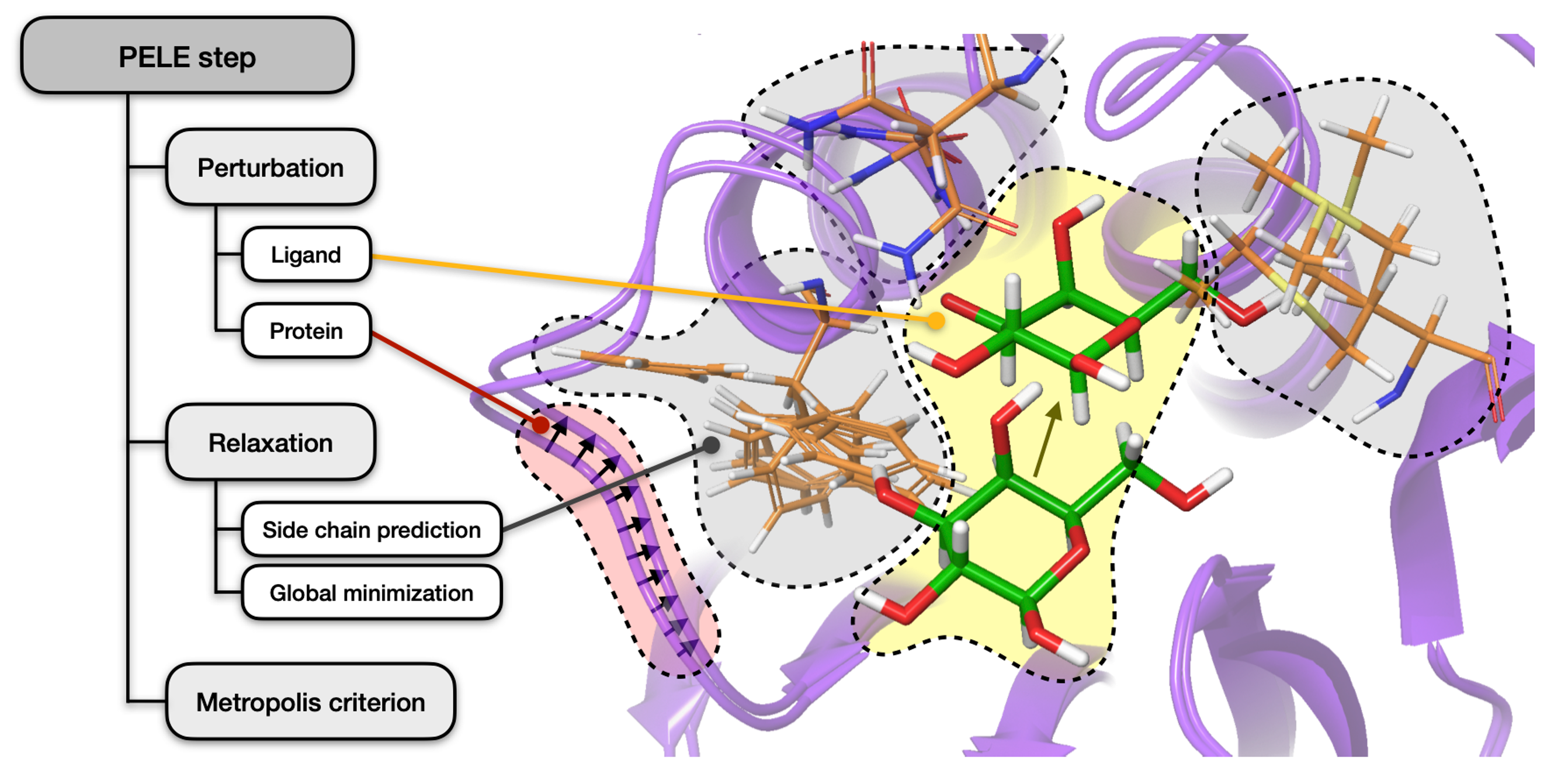
Stages of every PELE step¶
AdaptivePELE¶
AdaptivePELE is a Python package developed around the core PELE algorithm aimed to enhance the exploration of standard molecular simulations by iteratively running short simulations, assessing the exploration with a clustering, and spawning new trajectories in interesting regions.
The algorithm is composed of three main steps: sampling, clustering, and spawning, which run in an iterative approach.
1. Sampling. During this phase, several trajectories are run independently to generate a number of uncorrelated poses.
2. Clustering. Obtained conformations are clustered using the leader algorithm based on ligand RMSD.
3. Spawning. In the last step, the initial structures (seeds) for the next iteration are selected with the goal of improving the search in poorly sampled regions or to optimize a user-defined metric.
Check how to set it up here.
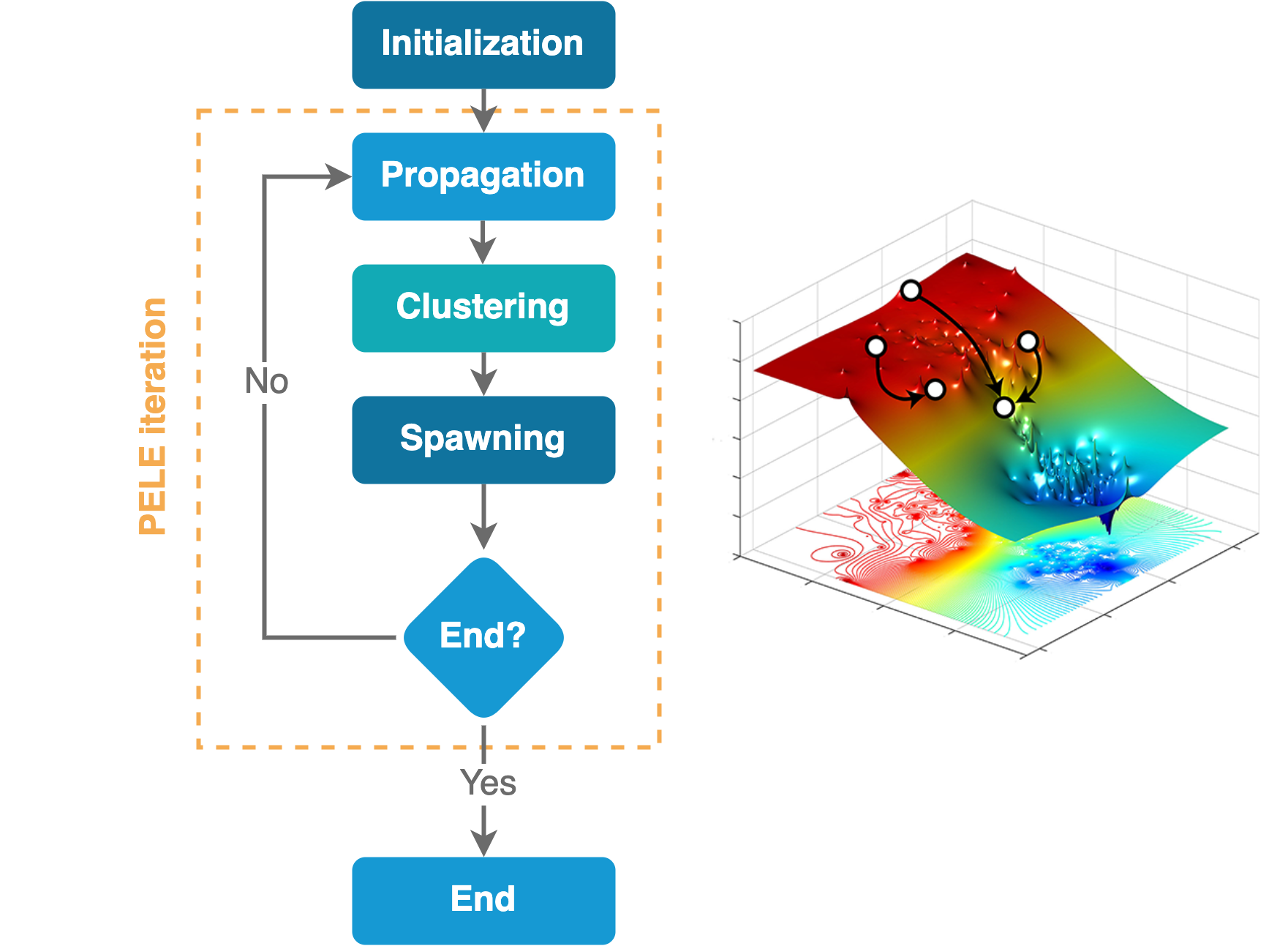
Flow chart depicting the AdaptivePELE algorithm¶
AquaPELE¶
AquaPELE extends the exploration capabilities of the standard PELE algorithm by introducing an additional Monte Carlo step to perturb water molecules inside protein cavities and dynamically adjust their effects to the current state of the system.
The implementation employs a mixed implicit/explicit approach which allows prediction of the principal hydration sites or the rearrangement and displacement of conserved water molecules upon the binding of a ligand while retaining the efficiency.
Check how to set it up here.
Conformation perturbation¶
PELE provides the possibility to narrow down the range of available ligand conformations to increase the efficiency of sampling. It will automatically generate a library of conformations when supplied with a directory of ligand clusters originating from conformational search or Bioactive Conformational Ensemble server.
Check how to set it up here.
FragPELE¶
FragPELE is a new tool for in silico hit-to-lead drug design, capable of growing a fragment into a core while exploring the protein-ligand conformational space.
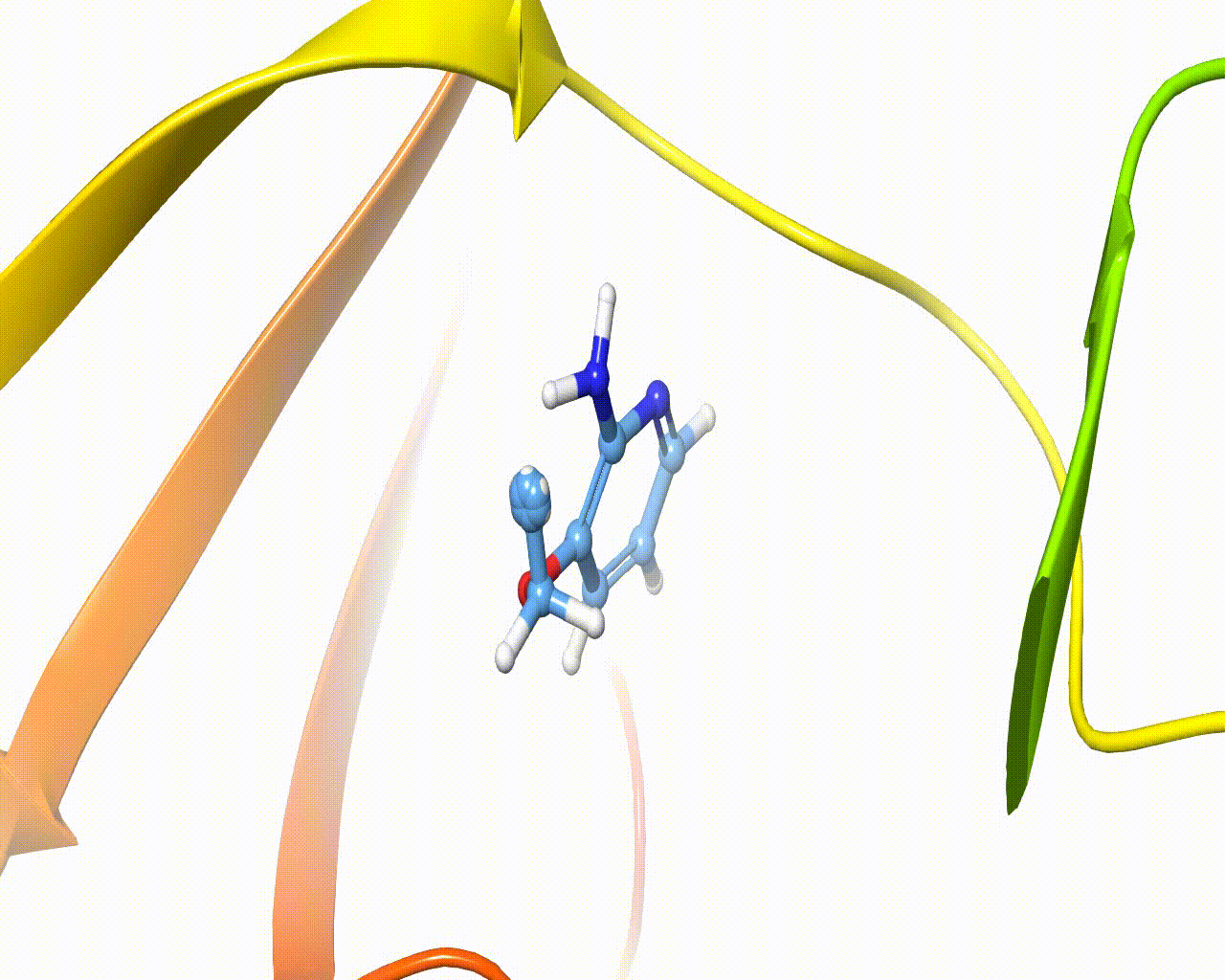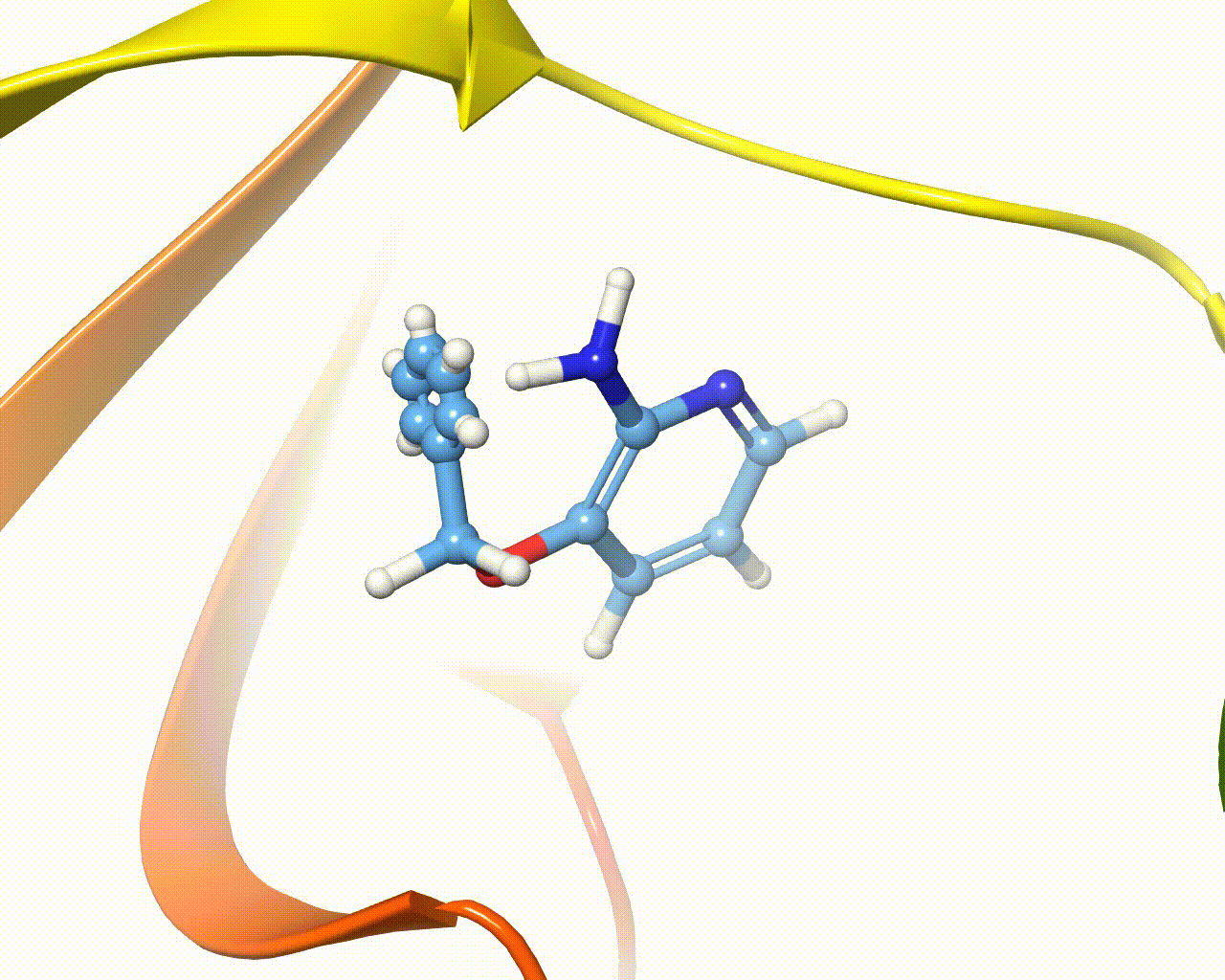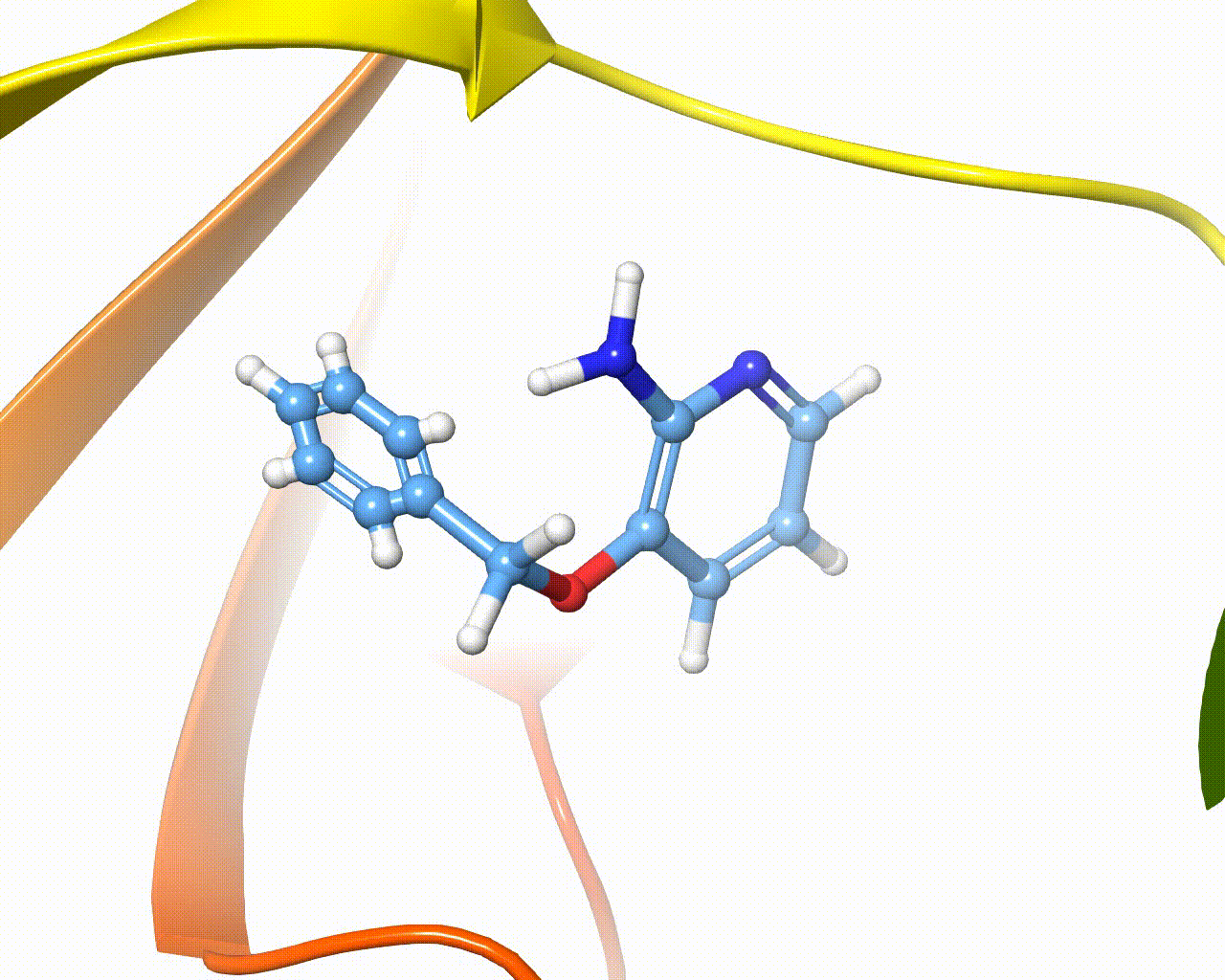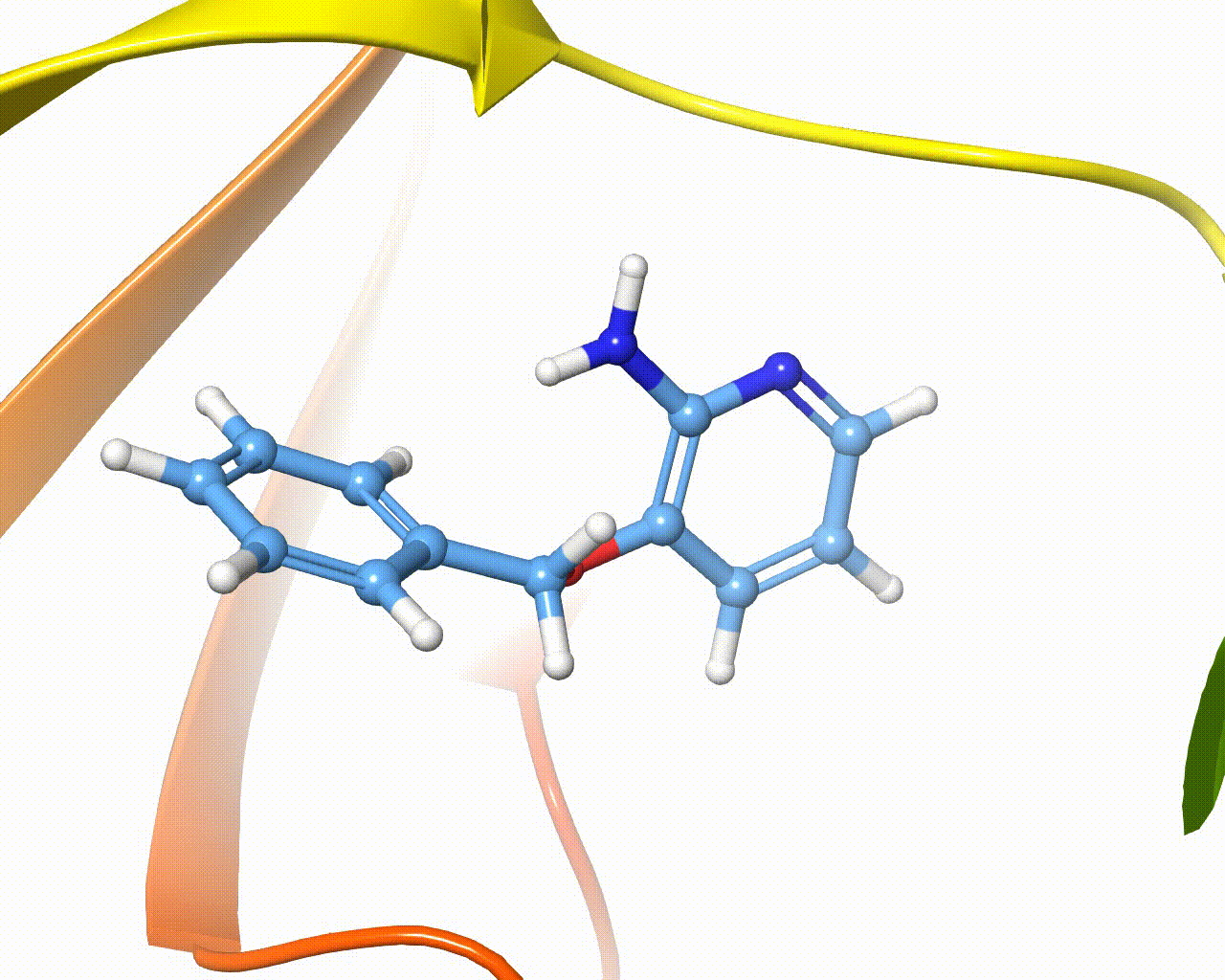
Visualization of fragment growing in a protein cavity¶
The software is intended to automatically grow one or more fragments onto different hydrogen atoms of the same scaffold, the overall method is composed of five steps, as outlined below:
1. Preparation. Preprocessing of the PDB file to ensure the protein and the scaffold are correctly protonated and have assigned bond orders.
2. Fragment linkage. In order to link the fragment to the scaffold, the coordinates of the hydrogens associated with user-defined heavy atoms will be aligned, and the hydrogens will be subsequently deleted to create a new bond.
3. Fragment reduction. The bonding and non-bonding terms of the fragment atoms are reduced to later be grown dynamically within the binding site.
4. Fragment growing. The new fragment is grown within a few epochs using the AdaptivePELE methodology. At each epoch, the atoms’ parameters are linearly increased and a few PELE steps are performed to account for the receptor flexibility.
5. Sampling & scoring. Once the ligand is completely grown, a longer PELE simulation is performed to score the and map the whole protein-ligand conformational space.
Check how to set it up here.
Further reading¶
Over the years, numerous publications have been written about the methodology and applications of PELE itself, as well as further improvements, such as AdaptivePELE or FragPELE.
PELE: Protein Energy Landscape Exploration. A Novel Monte Carlo Based Technique by Kenneth W. Borrelli, Andreas Vitalis, Raul Alcantara, and Victor Guallar
Adaptive simulations, towards interactive protein-ligand modeling by Daniel Lecina, Joan F. Gilabert, and Victor Guallar
aquaPELE: A Monte Carlo-Based Algorithm to Sample the Effects of Buried Water Molecules in Proteins by Martí Municoy, Sergi Roda, Daniel Soler, Alberto Soutullo, and Victor Guallar
Bioactive Conformational Ensemble Server and Database. A Public Framework to Speed Up In Silico Drug Discovery by Sanja Zivanovic, Genís Bayarri, Francesco Colizzi, David Moreno, Josep Lluís Gelpí, Robert Soliva, Adam Hospital and Modesto Orozco
FragPELE: Dynamic Ligand Growing within a Binding Site. A Novel Tool for Hit-To-Lead Drug Design by Carles Perez, Daniel Soler, Robert Soliva, and Victor Guallar