Input files preparation¶
PELE Platform essentially needs a minimum of two files in order to run:
system.pdb: a PDB file that contains the structure to simulate with PELE. It typically contains a protein and a small molecule.
input.yaml: a yaml file containing a collection of parameters to set up the simulation.
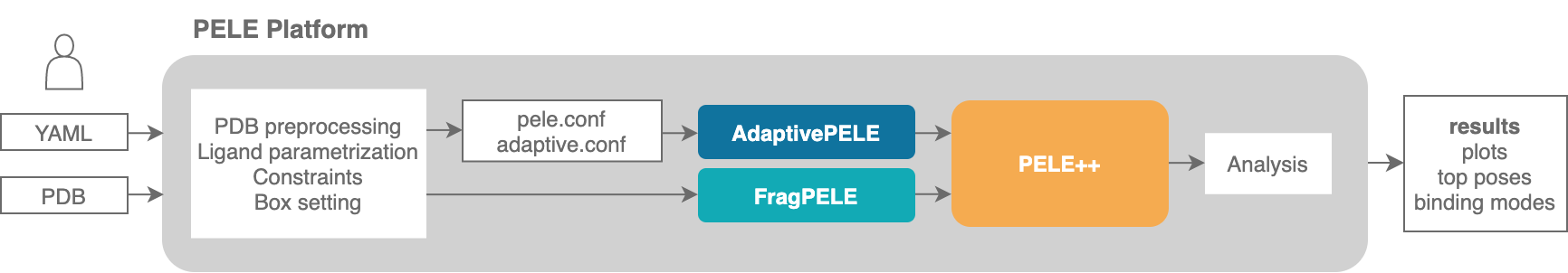
Additionally, although the Platform can create them automatically, we might want to supply custom templates for our ligand or other non standard residues.
templates: optionally, we can supply parameter templates or rotamer libraries for specific non standard residues.
Below we list several instructions to correctly prepare input PDB files for PELE:
On the other hand, the list of all parameters that can be specified in the
input.yaml file can be found below:
Finally, the instructions to manage custom templates are: